Georgia Tech Research Institute (GTRI) is looking into ways to speed up DNA-based cold storage in a $25m Scalable Molecular Archival Software and Hardware (SMASH) project.
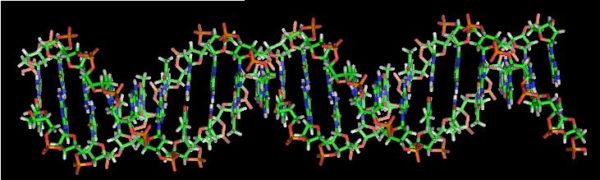
DNA is a biopolymer molecule composed from two chains in a double helix formation, and carrying genetic information. The chains are made up from nucleotides containing one of four nucleobases; cytosine (C), guanine (G), adenine (A) and thymine (T). Both chains carry the same data, which is encoded into sequences of the four nucleobases.

GTRI senior research scientist Nicholas Guise said in a quote that DNA storage: “is so compact that a practical DNA archive could store an exabyte of data, equivalent to a million terabyte hard drives, in a volume about the size of a sugar cube.”
Put another way, Alexa Harter, director of GTRI’s Cybersecurity, Information Protection, and Hardware Evaluation Research (CIPHER) Laboratory, said: “What would take acres in a data farm today could be kept in a device the size of the tabletop.”
The intent is to encode and decode terabytes of data in a day at costs and rates more than 100 times better than current technologies.
This is still slow by HDD and SSD standards. The intent is to use DNA storage for data that must be kept indefinitely, but accessed infrequently; backup/archive-type data in other words.
Guise said: “Scientists have been able to read DNA from animals that died centuries ago, so the data lasts essentially forever under the right conditions.”
The grant has been awarded by the Intelligence Advanced Research Projects Activity’s (IARPA) Molecular Information Storage (MIST) program and is for a multi-phase project involving;
- Georgia Tech’s Institute for Electronics and Nanotechnology – will provide fabrication facilities,
- Twist Bioscience – will engineer a DNA synthesis platform on silicon that “writes” the DNA strands which code the stored data,
- Roswell Biotechnologies – will provide molecular electronic DNA reader chips which are under development,
- The University of Washington, collaborating with Microsoft – will provide system architecture, data analysis and coding expertise.
GTRI envisages a hybrid chip with DNA grown above standard CMOS layers containing the electronics. Current technology uses modified inkjet printing to produce DNA strands. The SMASH project plans to grow the biopolymer more rapidly and in larger quantities using parallelized synthesis on these hybrid chips.

Data will be read from DNA strands using a molecular electronic sensor array chip, on which single molecules are drawn through nanoscale current meters that measure the electrical signatures of each letter, C, G, A and T, in the nucleotide sequence.
GTRI research engineer Brooke Becker said: “We’ll be working with commercial foundries, so when we get the processing right, it should be much easier to transition the technology over to them. Connecting to the existing technology infrastructure is a critical part of this project, but we’ll have to custom-make most of the components in the first stage.”
Guise cast more light on the difficulties: “The basic synthesis is proven at a scale of hundreds of microns. We want to shrink that by a factor of 100, which leads us to worry about such issues as crosstalk between different DNA strands in adjacent locations on the chips.”
Current human genome sequencing in biomedicine hopes to achieve a $1,000/genome cost. The SMASH project is looking for a $10/data genome cost. This is a huge difference; a hundredth less.
Blocks & Files thinks we’re looking at two to three year project here.
GTRI senior research scientist Adam Meier said: “We don’t see any killers ahead for this technology. There is a lot of emerging technology and doing this commercially will require many orders of magnitude improvement. Magnetic tape for archival storage has been improving steadily for 60 years, and this investment from IARPA will power the advancements needed to make DNA storage competitive with that.”
We could image a DNA helix as a kind of ribbon or tape, only at a molecular level. Storing an exabyte in a sugar cube-sized chip containing it would certainly make tape density look pretty shabby.