
Researchers at the University of Toronto (U of T) and NYU Grossman School of Medicine have developed a novel artificial intelligence technology, called ZFDesign, that can design zinc finger (ZF) proteins to target any stretch of DNA in the human genome, opening a door toward gene therapies for a broader range of health conditions. The researchers fed data from billions of interactions between ZF proteins and DNA into a machine-learning model, which can then generate engineered zinc fingers that bind to the given DNA sequence.
“Designing zinc fingers to bind specific DNA targets has been an unsolved problem for decades,” said Philip M. Kim, PhD, a professor in the Donnelly Centre for Cellular and Biomolecular Research at U of T’s Temerty Faculty of Medicine. “Our work should enable a new generation of in vivo therapeutics, which have proven difficult to develop with CRISPR and other DNA targeting technologies.”
David Ichikawa, PhD, a former graduate student at NYU Langone Health, added: “Our program can identify the right grouping of zinc fingers for any modification, making this type of gene editing faster than ever before.”
Ichikawa is lead author of the team’s published paper in Nature Biotechnology, which is titled “A universal deep-learning model for zinc finger design enables transcription factor reprogramming.” The investigators stated, “In this study, we present ZFDesign, a hierarchical attention-based artificial intelligence model trained on comprehensive screens of ZF–DNA interactions that consider the influence of multiple adjacent finger environments … ZFDesign captures these influences to provide a general design model for ZF arrays.”
Disorders including cystic fibrosis, Tay-Sachs disease, and sickle cell anemia are caused by errors in the order of DNA bases that encode the operating instructions for every human cell. Scientists can in some cases correct these mistakes with gene editing methods. Other conditions are caused not by a mistake in the DNA code itself, but by problems in how the cellular machinery reads DNA—epigenetics.
A gene often partners with transcription factors that tell the cell how much of that protein to make. When this process goes awry, over- or underactive genes can contribute to diseases including diabetes, cancer, and neurological disorders. As a result, researchers have been exploring ways to restore normal epigenetic activity. “Programmable regulation of gene expression would offer both powerful research tools as well as enormous therapeutic potential,” the authors stated. “Diseases caused by haploinsufficiency, gain-of-function mutations, or misexpression of a gene can be directly treated by modification of gene expression.”
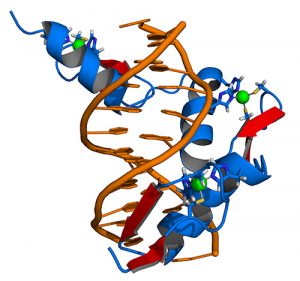
ZFs are a common class of human proteins that regulate such gene expression, a process that transcribes genetic information into RNA molecules and proteins. Scientists have long seen their potential because they naturally bind with DNA, are less likely than CRISPR and related technologies to trigger an immune reaction, and are small enough to work with clinical delivery methods. ZF editing can both change and control genes. Among the most abundant protein structures in the human body, ZFs can guide DNA repair by grabbing onto scissor-like enzymes and directing them to cut faulty segments out of the code.
Similarly, ZFs can also hook onto transcription factors and pull them toward a gene segment in need of regulation. By customizing these instructions, genetic engineers can tailor any gene’s activity.
A drawback, however, is that artificial ZFs are challenging to design for a specific task. “… the structurally intricate engagement of ZF domains with DNA has made their design challenging,” the authors pointed out. Since these proteins attach to DNA in complex groups, researchers would need to be able to tell—out of countless possible combinations—how every ZF interacts with its neighbor for each desired genetic change. So, for every new DNA target, scientists would have to engineer a new protein through a laborious and often unsuccessful workflow. The investigators noted, “While the potential utility of designer ZF arrays has long been recognized, their engineering has remained challenging with no proper design code having emerged thus far.”
This is not for lack of effort, they continued, because multiple approaches have been used to generate ZF libraries and ZF modules to provide designer ZF arrays. Even so, the team stated, “These approaches either require multiple rounds of laborious selection that produce ZFs with inconsistent activity or the application of preselected modules that often fail when expressed out of their selected context.”
The newly reported ZFDesign approach solves this problem with a universal model that offers ease of use comparable to CRISPR, and potentially higher DNA specificity. The technology uses artificial intelligence to model and design the interactions. “Because half of human TFs use ZFs to engage DNA, we reasoned that these endogenous ZF domains could be seamlessly replaced by designed ZFs without impacting the protein’s regulatory function,” the team commented. “This approach presents the designed ZFs in the exact context in which ZFs would occur naturally in the parent protein.”
“I think this system levels the playing field for zinc fingers and CRISPR,” said Kim, who is also a professor of molecular genetics and computer science at U of T. “CRISPR is very established for fundamental science, but our system has many advantages for applications in living systems, in particular, that zinc fingers are human proteins and would be safer as injected drugs.”
The ZFDesign technology can also generate many different proteins that do the same thing, offering more options for moving treatments toward the clinic. “Our program can identify the right grouping of zinc fingers for any modification, making this type of gene editing faster than ever before,” commented Ichikawa.
The ZF model has been developed with a research group at NYU Langone Health, led by Marcus Noyes, PhD, an assistant professor of biochemistry and molecular pharmacology in the Institute for Systems Genetics at the Grossman School of Medicine.
The model is based on data generated by the screen of billions of possible ZF-DNA interactions in the researchers’ labs. The Noyes lab has studied zinc fingers for years, and compiled data on 49 billion interactions among zinc finger proteins and DNA by creating high-throughput screens of several zinc finger libraries. Their approach combined two levels of data: interactions between single zinc fingers and DNA, and between each zinc finger and its neighboring finger. These zinc finger pair interactions influence DNA binding and hence gene expression.
The machine learning model developed by Kim and his group mirrors the data synthesis approach in the Noyes lab. “Our model is hierarchical, so it takes advantage of existing data from their first stage of screening—and a subset of their stage-two data—to develop predictions about which zinc fingers will be compatible with others in certain contexts,” said co-first author Osama Abdin, a doctoral student in Kim’s lab.
The model is based in part on technology that also forms the basis of ChatGPT, a software application developed by OpenAI that simulates human conversation. The model generates amino acid sequences for ZF proteins using large, highly detailed data sets and techniques similar to natural language processing.
The researchers showed the utility of the ZF system by reprogramming human transcription factors—ZF proteins that regulate the transcription of DNA to RNA. Working with professors Tim Hughes, PhD, and Mikko Taipale, PhD, in the Donnelly Centre, they tweaked the DNA binding targets of several transcription factors and programmed them to either activate or repress multiple genes. “We present a generalizable design method that allows for the seamless replacement of a TF natural DNA-binding domain to direct the TF to any target of interest,” the authors stated. “These RTFs [reprogrammed transcription factors] can produce activation and repression activities similar to CRISPR-based tools, establishing these proteins as attractive therapeutics comprising solely human components.”
Clinical application of reprogrammed transcription factors could target diseases caused by haploinsufficiency—deletion or inactivation of a copied gene, as in some cancers and the connective tissue disorders known as Ehlers-Danlos syndrome—or those linked to toxic genetic repeats, as in neurodegenerative conditions such as ALS, Parkinson’s, and Huntington’s disease.
Kim said the system already generates designs for ZF proteins with clinical potential, and that both his team in Toronto and the group in New York were surprised by how well it worked. The new system also holds promise for gene editing and other applications where CRISPR is useful, although its impact will likely be strongest in the area of reprogramming transcription factors, Kim said. The study authors add that besides posing a lower immune risk, the small size of zinc-finger tools may also provide more flexible gene therapy techniques compared with CRISPR by enabling more ways to deliver the tools to the right cells in patients.
“By speeding up zinc-finger design coupled with their smaller size, our system paves the way for using these proteins to control multiple genes at the same time,” said Noyes. “In the future, this approach may help correct diseases that have multiple genetic causes, such as heart disease, obesity, and many cases of autism.”
The developers further commented, “ZFDesign represents an important advance because the design of ZFs for any given target is now available at the push of a button and is open to the academic community for the study of a myriad of academic and therapeutic applications, with the advantages of small size and low immunogenicity.”
Noyes, an assistant professor in the department of biochemistry and molecular pharmacology at NYU Langone, cautioned that, while promising, ZFs can be difficult to control. Since they are not always specific to a single gene, some combinations can affect DNA sequences beyond a particular target, leading to unintended changes in genetic code. As a result, Noyes said the team next plans to refine its AI program so it can build more precise ZF groupings that only prompt the desired edit. Noyes is also a member of NYU Langone’s Institute for System Genetics.
The next step is to improve specificity in the system. “The current model is designed to optimize the engagement of a given zinc finger protein with its target, but has no built-in awareness of interactions to other targets,” said Kim. “Optimizing for specificity will entail modeling these other interactions.”
The researchers are well on their way to building a model that offers more specificity, Kim said. “There is a lot of excitement around this work in the fields of epigenetics and gene therapy, and I look forward to seeing what we can achieve with this technology.”
Noyes is a co-founder of TBG Therapeutics, which is developing methods to design ZFs and apply them to treatments for diseases with genetic components.